File remodeling tools¶
Remodeling refers to the process of transforming a tabular file
into a different form in order to disambiguate the
information or to facilitate a particular analysis.
The remodeling operations are specified in a JSON (.json) file,
giving a record of the transformations performed.
There are two types of remodeling operations: transformation and summarization. The transformation operations modify the tabular files, while summarization produces an auxiliary information file but leaves the tabular files unchanged.
The file remodeling tools can be applied to any tab-separated value (.tsv) file
but are particularly useful for restructuring files representing experimental events.
Please read the File remodeling quickstart
tutorials for an introduction and basic use of the tools.
The file remodeling tools can be applied to individual files using the HED online tools or to entire datasets using the remodel command-line interface either by calling Python scripts directly from the command line or by embedding calls in a Jupyter notebook. The tools are also available as HED RESTful services. The online tools are particularly useful for debugging.
This user’s guide contains the following topics:
Overview of remodeling¶
Remodeling consists of restructuring and/or extracting information from tab-separated value files based on a specified list of operations contained in a JSON file.
Internally, the remodeling operations represent the tabular file using a Pandas DataFrame.
Transformation operations¶
Transformation operations, shown schematically in the following figure, are designed to transform an incoming tabular file into a new DataFrame without modifying the incoming data.

Transformation operations are stateless and do not save any context information or affect future applications of the transformation.
Transformations, themselves, do not have any output and just return a new, transformed DataFrame. In other words, transformations do not operate in place on the incoming DataFrame, but rather, they create a new DataFrame containing the result.
Typically, the calling program is responsible for reading and saving the tabular file, so the user can choose whether to overwrite or create a new file.
See the remodeling tool program interface section for information on how to call the operations.
Summarization operations¶
Summarization operations do not modify the input DataFrame but rather extract and save information in an internally stored summary dictionary as shown schematically in the following figure.
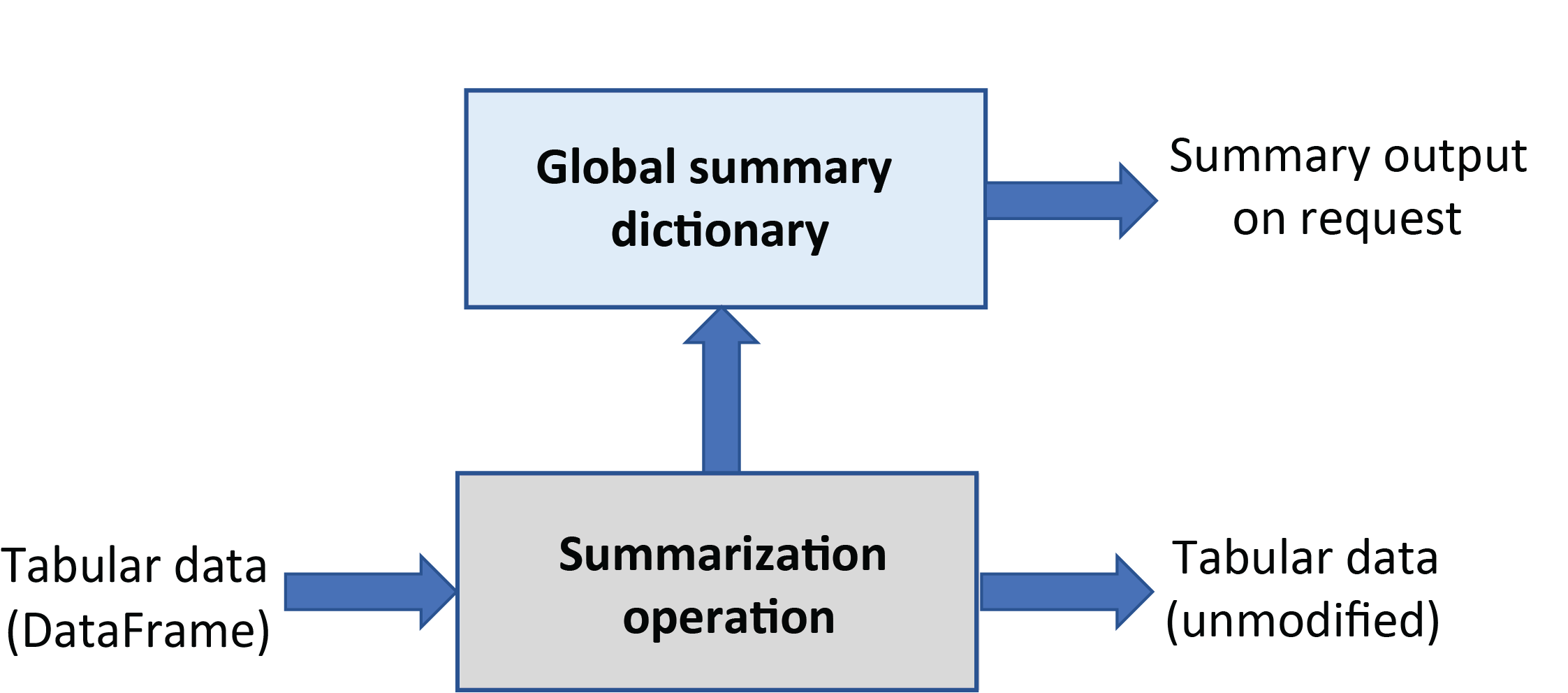
The dispatcher that executes remodeling operations can be interrogated at any time
for the state information contained in the global summary dictionary and can save additional summary information at any time during execution.
Usually summaries are dumped at the end of processing to the derivatives/remodel/summaries
subdirectory under the dataset root.
Summarization operations may appear anywhere in the operation list, and the same type of summary may appear multiple times under different names in order to track progress.
The dispatcher stores information from each uniquely named summarization operation as a separate summary dictionary entry. Within its summary information, most summarization operations keep a separate summary for each individual file and have methods to create an overall summary of the information for all the files that have been processed by the summarization.
Summarization results are available in JSON (.json) and text (.txt) formats.
Available operations¶
The following table lists the available remodeling operations with brief example use cases and links to further documentation. Operations not listed in the summarize section are transformations.
Category |
Operation |
Example use case |
|---|---|---|
clean-up |
||
Remove temporary columns created during restructuring. |
||
Remove rows with n/a values in a specified column. |
||
Make columns names consistent across a dataset. |
||
Make column order consistent across a dataset. |
||
factor |
||
Extract factor vectors from a column of condition variables. |
||
Extract factor vectors from search queries of HED annotations. |
||
Extract design matrices and/or condition variables. |
||
restructure |
||
Replace multiple consecutive events of the same type |
||
Create m columns from values in n columns (for recoding). |
||
Split trial-encoded rows into multiple events. |
||
summarize |
||
Summarize column names and order in the files. |
||
Count the occurrences of the unique column values. |
||
Summarize the HED tags present in the |
||
Summarize the detailed usage of a particular type tag |
||
Validate the data files and report any errors. |
||
Generate a sidecar template from an event file. |
The clean-up operations are used at various phases of restructuring to assure consistency across dataset files.
The factor operations produce column vectors with the same number of rows as the data file from which they were calculated. They encode condition variables, design matrices, or other search criteria. See the HED conditions and design matrices for more information on factoring and analysis.
The restructure operations modify the way in which a data file represents its information.
The summarize operations produce dataset-wide summaries of various aspects of the data files as well as summaries of the individual files.
Installing the remodel tools¶
The remodeling tools are available in the GitHub
hed-python repository
along with other tools for data cleaning and curation.
Although version 0.1.0 of this repository is available on PyPI
as hedtools, the version containing the restructuring tools (Version 0.2.0)
is still under development and has not been officially released.
However, the code is publicly available on the develop branch of the
hed-python repository and
can be directly installed from GitHub using pip:
pip install git+https://github.com/hed-standard/hed-python/@develop
The web services and online tools supporting remodeling are available
on the HED online tools dev server.
When version 0.2.0 of hedtools is officially released on PyPI, restructuring
will become available on the released HED online tools.
A docker version is also under development.
The following diagram shows a schematic of the remodeling process.
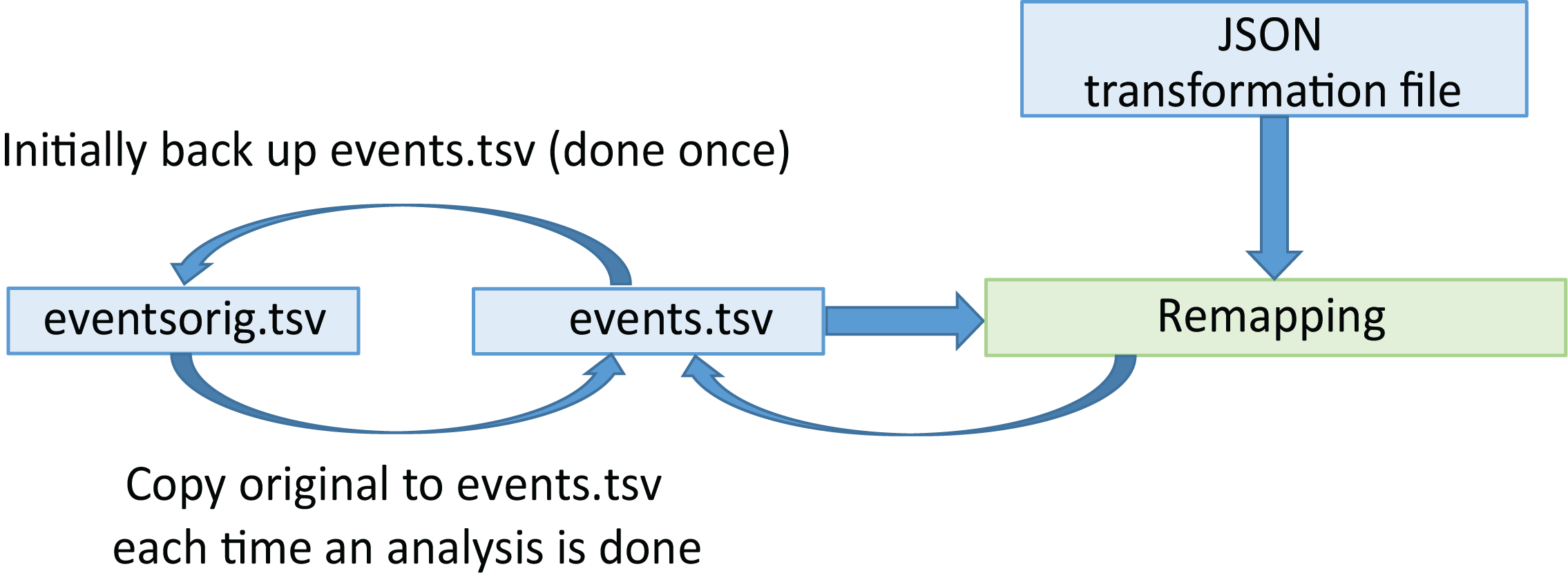
Initially, the user creates a backup of the specified tabular files (usually events.tsv files).
This backup is a mirror of the data files in the dataset,
but is located in the derivatives/remodel/backups directory and never modified once the backup is created.
Remodeling applies a sequence of operations specified in a JSON remodel file to the backup versions of the data files. The JSON remodel file provides a record of the operations performed on the file. If the user detects a mistake in the transformations, he/she can correct the transformation file and rerun the transformations.
Remodeling always runs on the original backup version of the file rather than the transformed version, so the transformations can always be corrected and rerun. It is possible to by-pass the backup, particularly if only using summarization operations, but this is not recommended and should be done with care.
Remodel command-line interface¶
The remodeling toolbox provides Python scripts with command-line interfaces to create or restore backups and to apply operations to the files in a dataset. The file remodeling tools may be applied to datasets that are in free form under a directory root or that are in BIDS-format. Some operations use HED (Hierarchical Event Descriptors) annotations. See the Remodel with HED section for a discussion of these operations and how to use them.
The remodeling command-line interface can be used from the command line, called from another Python program, or used in a Jupyter notebooks. Example notebooks can be found in the Jupyter notebooks to support remodeling.
Calling remodel tools¶
The remodeling tools provide three Python programs for backup (run_remodel_backup),
remodeling (run_remodel) and restoring (run_remodel_restore) event files.
These programs can be called from the command line or from another Python program.
The programs use a standard command-line argument list for specifying input as summarized in the following table.
Script name |
Arguments |
Purpose |
|---|---|---|
run_remodel_backup |
data_dir |
Create a backup event files. |
run_remodel |
data_dir |
Restructure or summarize the event files. |
run_remodel_restore |
data_dir |
Restore a backup of event files. |
All the scripts have a required argument, which is the full path of the dataset root (data_dir).
The run_remodel program has a required parameter which is the full path of a JSON file
containing a specification of the remodeling commands to be run.
Remodel command-line arguments¶
This section describes the arguments that are used for the remodeling command-line interface with examples and more details.
Positional arguments¶
Positional arguments are required and must be given in the order specified.
data_dir
The full path of dataset root directory.
model_path
The full path of the JSON remodel file (for run_remodel only).
Named arguments¶
Named arguments consist of a key starting with a hyphen and are possibly followed by a value. Named arguments can be given in any order or omitted. If omitted, a specified default is used. Argument keys and values are separated by spaces.
For argument values that are lists, the key is given followed by the items in the list, all separated by spaces.
Each command has two different forms of the key name: a short form (a single hyphen followed by a single character) and a longer form (two hyphens followed by a more self-explanatory name). Users are free to use either form.
-b, --bids-format
If this flag present, the dataset is in BIDS format with sidecars. Tabular files and their associated sidecars are located using BIDS naming.
-bd, --backup-dir
The path to the directory holding the backups (default:
[data_root]/derivatives/remodel/backups). Use the-nboption if you wish to omit the backup (inrun_remodel).
-bn, --backup-name
The name of the backup used for the remodeling (default:
default_back).
-e, --extensions
This option is followed by a list of file extension(s) of the data files to process. The default is
.tsv. Comma separated tabular files are not permitted.
-f, --file-suffix
This option is followed by the suffix names of the files to be processed. For example
events(the default) captures files namedevents.tsvif the default extension is used. The filename without the extension must end in one of the specified suffixes in order to be backed up or transformed.
-i, --individual-summaries
This option offers a choice among three options:
separate: Individual summaries for each file in separate files in addition the overall summary.
consolidated: Individual summaries written in the same file as the overall summary.
none: Only an overall summary.
-j, --json-sidecar
This option is followed by the full path of the JSON sidecar with HED annotations to be applied during the processing of HED-related remodeling operations.
-nb, --no-backup
If present, no backup is used. Rather operations are performed directly on the files.
-ns, --no-summaries
If present, no summary files are output.
-nu, --no-update
If present, the modified files are not output.
-r, --hed-versions
This option is followed by one or more HED versions. Versions of the standard schema are specified by their semantic versions (e.g.,
8.1.0), while library schema versions are prefixed by their library name (e.g.,score_1.0.0).
If more than one HED schema version is given, all but one of the versions must start with an additional namespace designator (e.g.,
sc:). At most one version can omit the namespace designator when multiple schema are being used. In annotations, tags must start with the namespace designator of the corresponding schema from which they were selected (e.g.sc:Sleep-modulatorif the SCORE library was designated bysc:score_1.0.0).
-s, --save-formats
This option is followed by the extensions (including .) of the formats in which to save summaries (default:
.txt.json).
-t, --task-names
The name(s) of the tasks to be included (for BIDS-formatted files only). When a dataset includes multiple tasks, the event files are often structured differently for each task and thus require different transformation files. This option allows the backups and operations to be restricted to an individual task.
If you omit this option, all tasks are used. This means that all
events.tsvfiles are restored from a backup if the backup is used, the operations are performed on allevents.tsvfiles, and summaries are combined over all tasks.
If a list of specific task names follows this option, only datafiles corresponding to the listed tasks are processed giving separate summaries for each listed task.
If a “*” follows this option, all event files are processed and separate summaries are created for each task.
Task detection follows the BIDS convention. Tasks are detected by finding “task-x” in the file names of
events.tsvfiles. Here x is the name of the task. The task name is followed by an underbar, by a period, or be at the end of the filename.
-v, --verbose
If present, more comprehensive messages documenting transformation progress are printed to standard output.
-w, --work-dir
The path to the remodeling work root directory –both for summaries (default:
[data_root]/derivatives/remodel). Use the-nboption if you wish to omit the backup (inrun_remodel).
-x, --exclude-dirs
The directories to exclude when gathering the data files to process. For BIDS datasets, these are typically
derivatives,stimuli, andsourcecode. Any subdirectory with a path component namedremodelis automatically excluded from remodeling, as these directories are reserved for storing backup, state, and result information for the remodeling process itself.
Remodel scripts¶
This section discusses the three main remodeling scripts with command-line interfaces to support backup, remodeling, and restoring the tabular files used in the remodeling process. These scripts can be run from the command line or from another Python program using a function call.
Backing up files¶
The run_remodel_backup Python program creates a backup of the specified files.
The backup is always created in the derivatives/remodel/backups subdirectory
under the dataset root as shown in the following example for the
sample dataset eeg_ds003645s_hed_remodel,
which can be found in the datasets subdirectory of the
hed-examples GitHub repository.
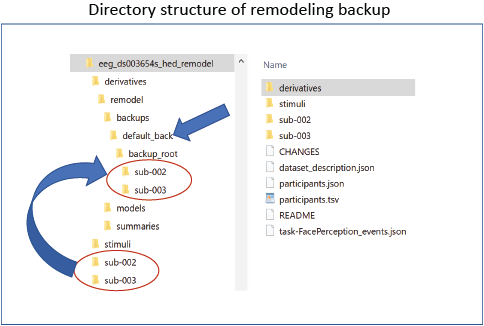
The backup process creates a mirror of the directory structure of the source files to be backed up
in the directory derivatives/remodel/backups/backup_name/backup_root as shown in the figure above.
The default backup name is default_back.
In the above example, the backup has subdirectories sub-002 and sub-003 just
like the main directory of the dataset.
These subdirectories only contain backups of the files to be transformed
(by default files with names ending in events.tsv).
In addition to the backup_root, the backup directory also contains a dictionary of backup files
in the backup_lock.json file. This dictionary is used internally by the remodeling tools.
The backup should be created once and not modified by the user.
The following example shows how to run the run_remodel_backup program from the command line
to back up the dataset located at /datasets/eeg_ds003645s_hed_remodel.
Example of calling run_remodel_backup from the command line.
python run_remodel_backup /datasets/eeg_ds003645s_hed_remodel -x derivatives stimuli
Since the -f and -e arguments are not given, the default file suffix and extension values
apply, so only files of the form events.tsv are backed up.
The -x option excludes any source files from the derivatives and stimuli subdirectories.
These choices can be overridden using additional command-line arguments.
The following shows how the run_remodel_backup program can be called from a
Python program or a Jupyter notebook.
The command-line arguments are given in a list instead of on the command line.
Example of Python code to call run_remodel_backup using a function call.
import hed.tools.remodeling.cli.run_remodel_backup as cli_backup
data_root = '/datasets/eeg_ds003645s_hed_remodel'
arg_list = [data_root, '-x', 'derivatives', 'stimuli']
cli_backup.main(arg_list)
During remodeling, each file in the source is associated with a backup file using its relative path from the dataset root. Remodeling is performed by reading the backup file, performing the operations specified in the JSON remodel file, and overwriting the source file as needed.
Users can also create alternatively named backups by providing the -n argument with a backup name to
the run_remodel_backup program.
To use backup files from another named backup, call the remodeling program with
the -n argument and the correct backup name.
Named backups can provide checkpoints to allow the execution of
transformations to start from intermediate points.
NOTE: You should not delete backups, even if you have created multiple named backups. The backups provide useful state and provenance information about the data.
Remodeling files¶
Remodeling consists of applying a sequence of operations from the remodel operation summary to successively transform each backup file according to the instructions and to overwrite the actual files with the final result.
If the dataset has no backups, the actual data files rather than the backups are transformed. You are expected to create the backup (just once) before running the remodeling operations. Going without backup is not recommended unless you are only doing summarization operations.
The operations are specified as a list of dictionaries in a JSON file in the remodel sample files as discussed below.
Before running remodeling transformations on an entire dataset, consider using the HED online tools to debug your remodeling operation file on a single file. The remodeling process always starts with the original backup files, so the usual development path is to incrementally add operations to the end of your transformation JSON file as you develop and test on a single file until you have the desired end result.
The following example shows how to run a remodeling script from the command line. The example assumes that the backup has already been created for the dataset.
Example of calling run_remodel from the command line.
python run_remodel /datasets/eeg_ds003645s_hed_remodel /datasets/remove_extra_rmdl.json -x derivatives simuli
The script has two required arguments the dataset root and the path to the JSON remodel file.
Usually, the JSON remodel files are stored with the dataset itself in the
derivatives/remodel/remodeling_files subdirectory, but common scripts can be stored in a central place elsewhere.
The additional keyword option, -x in the example indicates that directory paths containing the component derivatives or the component stimuli should be excluded.
Excluded directories need not have their excluded path component at the top level of the dataset.
Subdirectory paths containing the remodel path component are automatically excluded.
The command-line interface can also be used in a Jupyter notebook or as part of a larger Python
program by calling the main function with the equivalent command-line arguments provided
in a list with the positional arguments appearing first.
The following example shows Python code to remodel a dataset using the command-line interface. This code can be used in a Jupyter notebook or in another Python program.
Example Python code to call run_remodel using a function call.
import hed.tools.remodeling.cli.run_remodel as cli_remodel
data_root = '/datasets/eeg_ds003645s_hed_remodel'
model_path = '/datasets/remove_extra_rmdl.json'
arg_list = [data_root, model_path, '-x', 'derivatives', 'stimuli']
cli_remodel.main(arg_list)
Restoring files¶
Since remodeling always uses the backed up version of each data file, there is no need to restore these files to their original state between remodeling runs. However, when finished with an analysis, you may want to restore the data files to their original state.
The following example shows how to call run_remodel_restore to
restore the data files from the default backup.
The restore operation restores all the files in the specified backup.
Example of calling run_remodel_restore from the command line.
python run_remodel_restore /datasets/eeg_ds003645s_hed_remodel
As with the other command-line programs, run_remodel_restore can be also called using a function call.
Example Python code to call run_remodel_restore using a function call.
import hed.tools.remodeling.cli.run_restore as cli_remodel
data_root = '/datasets/eeg_ds003645s_hed_remodel'
cli_remodel.main([data_root])
Remodel with HED¶
HED (Hierarchical Event Descriptors) is a system for annotating data in a manner that is both human-understandable and machine-actionable. HED provides much more detail about the events and their meanings, If you are new to HED see the HED annotation quickstart. For information about HED’s integration into BIDS (Brain Imaging Data Structure) see BIDS annotation quickstart.
Currently, five remodeling operations rely on HED annotations:
HED tags provide a mechanism for advanced data analysis and for extracting experiment-specific information from the data files. However, since HED information is not always stored in the data files themselves, you may need to provide a HED schema and a JSON sidecar.
The HED schema defines the allowed HED tag vocabulary, and the JSON sidecar associates HED annotations with the information in the columns of the event files. If you are not using any of the HED operations in your remodeling, you do not have to provide this information.
Extracting HED information from BIDS¶
The simplest way to use HED with run_remodel is to use the -b option,
which indicates that the dataset is in BIDS (Brain Imaging Data Structure) format.
BIDS is a standardized way of organizing neuroimaging data. HED and BIDS are well integrated. If you are new to BIDS, see the BIDS annotation quickstart.
A HED-annotated BIDS dataset provides the HED schema version in the dataset_description.json
file located directly under the BIDS dataset root.
BIDS datasets must have filenames in a specific format, and the HED tools can locate the relevant JSON sidecars for each data file based on this information.
Directly specifying HED information¶
If your data is already in BIDS format, using the -b option is ideal since
the needed information can be located automatically.
However, early in the experimental process,
your datafiles are not likely to be organized in BIDS format,
and this option will not be available if you want to use HED.
Without the -b option, the remodeling tools locate the appropriate files based
on specified filename suffixes and extensions.
In order to use HED operations, you must explicitly specify the HED versions
using the -r option.
The -r option supports a list of HED versions if multiple HED schemas are used.
For example: -r 8.1.0 sc:score_1.0.0 specifies that vocabulary will be drawn
from standard HED Version 8.1.0 and from
HED SCORE library version 1.0.0.
Annotations containing tags from SCORE should be prefixed with sc:.
Note: both of the schemas can be viewed by the HED Schema Viewer.
Usually, annotators will consolidate HED annotations in a single JSON sidecar file
located at the top-level of the dataset.
The path of this sidecar can be passed as a command-line argument using the -j option.
If more than one JSON sidecar file contains HED annotations, users will need to call the lower-level
remodeling functions to perform these operations.
The following example illustrates a command-line call that passes both a HED schema version and the path to the JSON file with the HED annotations.
Remodeling a non-BIDS dataset using HED.
python run_remodel /datasets/eeg_ds003645s_hed_remodel /datasets/summarize_conditions_rmdl.json \
-x derivatives simuli -r 8.1.0 -j /datasets/eeg_ds003645s_hed_remodel/task-FacePerception_events.json
Example Python code to use run_remodel on a non-BIDS dataset.
import hed.tools.remodeling.cli.run_remodel as cli_remodel
data_root = '/datasets/eeg_ds003645s_hed_remodel'
model_path = '/datasets/summarize_conditions_rmdl.json'
json_path = '/datasets/eeg_ds003645s_hed_remodel/task-FacePerception_events.json'
arg_list = [data_root, model_path, '-x', 'derivatives', 'stimuli', '-r' 8.1.0 '-j' json_path]
cli_remodel.main(arg_list)
Remodel error handling¶
Errors can occur during several stages in during remodeling and how they are handled depends on the type of error and where the error occurs. Except for the validation summary, the underlying remodeling code raises exceptions for most errors.
Errors in the remodel file¶
Each individual operation raises an exception if required parameters are missing or
the values provided for the parameters are of the wrong type.
However, the higher-level calling mechanisms provided through run_remodel
call the parse_operations static method provided by the Dispatcher to create
a parsed operation list.
This call either returns a list of parsed operations or a list of parse errors for
the operations in the list.
If there are any errors in the remodel file, no operations are run, but the errors for all operations in the list are reported. This allows users to correct errors in all operations in one pass without any data modification. The HED online tools are particularly useful for debugging the syntax and other issues in the remodeling process.
Execution-time remodel errors¶
When an error occurs during execution, an exception is raised. Exceptions are raised for invalid or missing files or if a transformed file is unable to be rewritten due to improper file permissions. Each individual operation may also raise an exception if the data file being processed does not have the expected information, such as a column with a particular name.
Exceptions raised during execution cause the process to be terminated and no further files are processed.
Remodel sample files¶
All remodeling operations are specified in a standardized JSON remodel input file.
The following shows the contents of the JSON remodeling file remove_extra_rmdl.json,
which contains a single operation with instructions to remove the value and sample columns
from the data file if the columns exist.
Sample remodel file¶
A sample JSON remodeling file with a single remove_columns transformation operation.
[
{
"operation": "remove_columns",
"description": "Remove unwanted columns prior to analysis",
"parameters": {
"remove_names": ["value", "sample"],
"ignore_missing": true
}
}
]
Each operation is specified in a dictionary with three top-level keys: “operation”, “description”, and “parameters”. The value of the “operation” is the name of the operation. The “description” value should include the reason this operation was needed, not just a description of the operation itself. Finally, the “parameters” value is a dictionary mapping parameter name to parameter value.
The parameters for each operation are listed in Remodel transformations and Remodel summarizations sections. An operation may have both required and optional parameters. Optional parameters may be omitted if unneeded, but all parameters are specified in the “parameters” section of the dictionary.
The remodeling JSON files should have names ending in _rmdl.json to more easily
distinguish them from other JSON files.
Although these files can be stored anywhere, their preferred location is
in the deriviatves/remodel/models subdirectory under the dataset root so
that they can provide provenance for the dataset.
Sample remodel event file¶
Several examples illustrating the remodeling operations use the following excerpt of the stop-go task from sub-0013 of the AOMIC-PIOP2 dataset available on OpenNeuro as ds002790. The full event file is sub-0013_task-stopsignal_acq-seq_events.tsv.
Excerpt from an event file from the stop-go task of AOMIC-PIOP2 (ds002790).
onset |
duration |
trial_type |
stop_signal_delay |
response_time |
response_accuracy |
response_hand |
sex |
|---|---|---|---|---|---|---|---|
0.0776 |
0.5083 |
go |
n/a |
0.565 |
correct |
right |
|
5.5774 |
0.5083 |
unsuccesful_stop |
0.2 |
0.49 |
correct |
right |
female |
9.5856 |
0.5084 |
go |
n/a |
0.45 |
correct |
right |
female |
13.5939 |
0.5083 |
succesful_stop |
0.2 |
n/a |
n/a |
n/a |
female |
17.1021 |
0.5083 |
unsuccesful_stop |
0.25 |
0.633 |
correct |
left |
male |
21.6103 |
0.5083 |
go |
n/a |
0.443 |
correct |
left |
male |
Sample remodel sidecar file¶
For remodeling operations that use HED, a JSON sidecar is usually required to provide the necessary HED annotations. The following JSON sidecar excerpt is used in several examples to illustrate some of these operations. The full JSON file can be found at task-stopsiqnal_acq-seq_events.json.
Excerpt of JSON sidecar with HED annotations for the stop-go task of AOMIC-PIOP2.
{
"trial_type": {
"HED": {
"succesful_stop": "Sensory-presentation, Visual-presentation, Correct-action, Image, Label/succesful_stop",
"unsuccesful_stop": "Sensory-presentation, Visual-presentation, Incorrect-action, Image, Label/unsuccesful_stop",
"go": "Sensory-presentation, Visual-presentation, Image, Label/go"
}
},
"stop_signal_delay": {
"HED": "(Auditory-presentation, Delay/# s)"
},
"sex": {
"HED": {
"male": "Def/Male-image-cond",
"female": "Def/Female-image-cond"
}
},
"hed_defs": {
"HED": {
"def_male": "(Definition/Male-image-cond, (Condition-variable/Image-sex, (Male, (Image, Face))))",
"def_female": "(Definition/Female-image-cond, (Condition-variable/Image-sex, (Female, (Image, Face))))"
}
}
}
Notice that the JSON file has some keys (e.g., “trial_type”, “stop_signal_delay”, and “sex”) which also correspond to columns in the events file. The “hed_defs” key corresponds to an extra entry in the JSON file that, in this case, provides the definitions needed in the HED annotation.
HED operations also require the HED schema. Most of the examples use HED standard schema version 8.1.0.
Remodel transformations¶
Factor column¶
The factor_column operation appends factor vectors to tabular files based on the values in a specified file column. Each factor vector contains a 1 if the corresponding row had that column value and a 0 otherwise. The factor_column is used to reformat event files for analyses such as linear regression based on column values.
Factor column parameters¶
Parameters for the factor_column operation.
Parameter |
Type |
Description |
|---|---|---|
column_name |
str |
The name of the column to be factored. |
factor_values |
list |
Column values to be included as factors. |
factor_names |
list |
Column names for created factors. |
If column_name is not a column in the data file, a ValueError is raised.
If factor_values is empty, factors are created for each unique value in column_name. Otherwise, only factors for the specified column values are generated. If a specified value is missing in a particular file, the corresponding factor column contains all zeros.
If factor_names is empty, the newly created columns are of the form column_name.factor_value. Otherwise, the newly created columns have names factor_names. If factor_names is not empty, then factor_values must also be specified and both lists must be of the same length.
Factor column example¶
The factor_column operation in the following example specifies that factor columns should be created for succesful_stop and unsuccesful_stop of the trial_type column. The resulting columns are called stopped and stop_failed, respectively.
A sample JSON file with a single factor_column transformation operation.
[{
"operation": "factor_column",
"description": "Create factors for the succesful_stop and unsuccesful_stop values.",
"parameters": {
"column_name": "trial_type",
"factor_values": ["succesful_stop", "unsuccesful_stop"],
"factor_names": ["stopped", "stop_failed"]
}
}]
The results of executing this factor_column operation on the sample remodel event file are:
Results of the factor_column operation on the sampe data.
onset |
duration |
trial_type |
stop_signal_delay |
response_time |
response_accuracy |
response_hand |
sex |
stopped |
stop_failed |
|---|---|---|---|---|---|---|---|---|---|
0.0776 |
0.5083 |
go |
n/a |
0.565 |
correct |
right |
female |
0 |
0 |
5.5774 |
0.5083 |
unsuccesful_stop |
0.2 |
0.49 |
correct |
right |
female |
0 |
1 |
9.5856 |
0.5084 |
go |
n/a |
0.45 |
correct |
right |
female |
0 |
0 |
13.5939 |
0.5083 |
succesful_stop |
0.2 |
n/a |
n/a |
n/a |
female |
1 |
0 |
17.1021 |
0.5083 |
unsuccesful_stop |
0.25 |
0.633 |
correct |
left |
male |
0 |
1 |
21.6103 |
0.5083 |
go |
n/a |
0.443 |
correct |
left |
male |
0 |
0 |
Factor HED type¶
The factor_hed_type operation produces factor columns based on values of the specified HED type tag. The most common type is the HED Condition-variable tag, which corresponds to factor vectors based on the experimental design. Other commonly use type tags include Task, Control-variable, and Time-block.
We assume that the dataset has been annotated using HED tags to properly document information such as experimental conditions, and focus on how such an annotated dataset can be used with remodeling to produce factor columns corresponding to these type variables.
For additional information on how to encode experimental designs using HED, see HED conditions and design matrices.
Factor HED type parameters¶
Parameters for factor_hed_type operation.
Parameter |
Type |
Description |
|---|---|---|
type_tag |
str |
HED tag used to find the factors (most commonly Condition-variable). |
type_values |
list |
Values to factor for the type_tag. |
Factor HED type example¶
The factor_hed_type operation in the following example appends additional columns to each data file corresponding to each possible value of each Condition-variable tag. The columns contain 1’s for rows corresponding to rows (e.g., events) for which that condition applies and 0’s otherwise.
A JSON file with a single factor_hed_type transformation operation.
[{
"operation": "factor_hed_type",
"description": "Factor based on the sex of the images being presented.",
"parameters": {
"type_tag": "Condition-variable",
"type_values": []
}
}]
The results of executing this factor_hed-tags operation on the sample remodel event file using the sample remodel sidecar file for HED annotations are:
Results of factor_hed_type.
onset |
duration |
trial_type |
stop_signal_delay |
response_time |
response_accuracy |
response_hand |
sex |
Image-sex.Female-image-cond |
Image-sex.Male-image-cond |
|---|---|---|---|---|---|---|---|---|---|
0.0776 |
0.5083 |
go |
n/a |
0.565 |
correct |
right |
female |
1 |
0 |
5.5774 |
0.5083 |
unsuccesful_stop |
0.2 |
0.49 |
correct |
right |
female |
1 |
0 |
9.5856 |
0.5084 |
go |
n/a |
0.45 |
correct |
right |
female |
1 |
0 |
13.5939 |
0.5083 |
succesful_stop |
0.2 |
n/a |
n/a |
n/a |
female |
1 |
0 |
17.1021 |
0.5083 |
unsuccesful_stop |
0.25 |
0.633 |
correct |
left |
male |
0 |
1 |
21.6103 |
0.5083 |
go |
n/a |
0.443 |
correct |
left |
male |
0 |
1 |
Merge consecutive¶
Sometimes a single long event in experimental logs is represented by multiple repeat events. The merge_consecutive operation collapses these consecutive repeat events into one event with duration updated to encompass the temporal extent of the merged events.
Merge consecutive parameters¶
Parameters for the merge_consecutive operation.
Parameter |
Type |
Description |
|---|---|---|
column_name |
str |
The name of the column which is the basis of the merge. |
event_code |
str, int, float |
The value in column_name that triggers the merge. |
match_columns |
list |
Columns whose values must match to collapse events. |
set_durations |
bool |
If true, set durations based on merged events. |
ignore_missing |
bool |
If true, missing column_name or match_columns do not raise an error. |
The first of the group of rows (each representing an event) to be merged is called the anchor
for the merge. After the merge, it is the only row in the group
that remains in the data file. The result is identical
to its original version, except for the value in the duration column.
If the set_duration parameter is true, the new duration is calculated as though
the event began with the onset of the first event (the anchor row) in the group and
ended at the point where all the events in the group have ended.
This method allows for small gaps between events and for events in which an
intermediate event in the group ends after later events.
If the set_duration parameter is false, the duration of the merged row is set to n/a.
If the data file has other columns besides onset, duration and column column_name,
the values in the other columns must be considered during the merging process.
The match_columns is a list of the other columns whose values must agree with those
of the anchor row in order for a merge to occur. If match_columns is empty, the
other columns in each row are not taken into account during the merge.
Merge consecutive example¶
The merge_consecutive operation in the following example causes consecutive
succesful_stop events whose stop_signal_delay, response_hand, and sex columns
have the same values to be merged into a single event.
A JSON file with a single merge_consecutive transformation operation.
[{
"operation": "merge_consecutive",
"description": "Merge consecutive *succesful_stop* events that match the *match_columns.",
"parameters": {
"column_name": "trial_type",
"event_code": "succesful_stop",
"match_columns": ["stop_signal_delay", "response_hand", "sex"],
"set_durations": true,
"ignore_missing": true
}
}]
When this operation is applied to the following input file,
the three events with a value of succesful_stop in the trial_type column starting
at onset value 13.5939 are merged into a single event.
Input file for a merge_consecutive operation.
onset |
duration |
trial_type |
stop_signal_delay |
response_hand |
sex |
|---|---|---|---|---|---|
0.0776 |
0.5083 |
go |
n/a |
right |
female |
5.5774 |
0.5083 |
unsuccesful_stop |
0.2 |
right |
female |
9.5856 |
0.5084 |
go |
n/a |
right |
female |
13.5939 |
0.5083 |
succesful_stop |
0.2 |
n/a |
female |
14.2 |
0.5083 |
succesful_stop |
0.2 |
n/a |
female |
15.3 |
0.7083 |
succesful_stop |
0.2 |
n/a |
female |
17.3 |
0.5083 |
unsuccesful_stop |
0.25 |
n/a |
female |
19.0 |
0.5083 |
unsuccesful_stop |
0.25 |
n/a |
female |
21.1021 |
0.5083 |
unsuccesful_stop |
0.25 |
left |
male |
22.6103 |
0.5083 |
go |
n/a |
left |
male |
Notice that the succesful_stop event at onset value 17.3 is not
merged because the stop_signal_delay column value does not match the value in the previous event.
The final result has duration computed as 2.4144 = 15.3 + 0.7083 - 13.5939.
The results of the merge_consecutive operation.
onset |
duration |
trial_type |
stop_signal_delay |
response_hand |
sex |
|---|---|---|---|---|---|
0.0776 |
0.5083 |
go |
n/a |
right |
female |
5.5774 |
0.5083 |
unsuccesful_stop |
0.2 |
right |
female |
9.5856 |
0.5084 |
go |
n/a |
right |
female |
13.5939 |
2.4144 |
succesful_stop |
0.2 |
n/a |
female |
17.3 |
2.2083 |
unsuccesful_stop |
0.25 |
n/a |
female |
21.1021 |
0.5083 |
unsuccesful_stop |
0.25 |
left |
male |
22.6103 |
0.5083 |
go |
n/a |
left |
male |
The events that had onsets at 17.3 and 19.0 are also merged in this example
Remap columns¶
The remap_columns operation maps combinations of values in m specified columns of a data file into values in n columns using a defined mapping. Remapping is useful during analysis to create columns in event files that are more directly useful or informative for a particular analysis.
Remapping is also important during the initial generation of event files from experimental logs. The log files generated by experimental control software often generate a code for each type of log entry. Remapping can be used to convert the column containing these codes into one or more columns with more informative information.
Remap columns parameters¶
Parameters for the remap_columns operation.
Parameter |
Type |
Description |
|---|---|---|
source_columns |
list |
A list of m names of the source columns for the map. |
destination_columns |
list |
A list of n names of the destination columns for the map. |
map_list |
list |
A list of mappings. Each element is a list of m source |
ignore_missing |
bool |
If false, source column values not in the map generate “n/a” |
integer_sources |
list |
[Optional] A list of source columns that are integers. |
A column cannot be both a source and a destination, and all source columns must be present in the data files. New columns are created for destination columns that are missing from a data file.
The remap_columns operation only works for columns containing strings or integers,
as it is meant for remapping categorical codes.
You must specify the which source columns contain integers so that n/a values
can be handled appropriately.
The map_list parameter specifies how each unique combination of values from the source columns will be mapped into the destination columns. If there are m source columns and n destination columns, then each entry in map_list must be a list with m + n elements. The first m elements are the key values from the source columns. The map_list should have targets for all combinations of values that appear in the m source columns unless ignore_missing is true.
After remapping, the tabular file will contain both source and destination columns. If you wish to replace the source columns with the destination columns, use a remove_columns transformation after the remap_columns.
Remap columns example¶
The remap_columns operation in the following example creates a new column called response_type based on the unique values in the combination of columns response_accuracy and response_hand.
A JSON file with a single remap_columns transformation operation.
[{
"operation": "remap_columns",
"description": "Map response_accuracy and response hand into a single column.",
"parameters": {
"source_columns": ["response_accuracy", "response_hand"],
"destination_columns": ["response_type"],
"map_list": [["correct", "left", "correct_left"],
["correct", "right", "correct_right"],
["incorrect", "left", "incorrect_left"],
["incorrect", "right", "incorrect_left"],
["n/a", "n/a", "n/a"]],
"ignore_missing": true
}
}]
In this example there are two source columns and one destination column, so each entry in map_list must be a list with three elements two source values and one destination value). Since all the values in map_list are strings, the optional integer_sources list is not needed.
The results of executing the previous remap_column command on the sample remodel event file are:
Mapping columns response_accuracy and response_hand into a response_type column.
onset |
duration |
trial_type |
stop_signal_delay |
response_time |
response_accuracy |
response_hand |
sex |
response_type |
|---|---|---|---|---|---|---|---|---|
0.0776 |
0.5083 |
go |
n/a |
0.565 |
correct |
right |
female |
correct_right |
5.5774 |
0.5083 |
unsuccesful_stop |
0.2 |
0.49 |
correct |
right |
female |
correct_right |
9.5856 |
0.5084 |
go |
n/a |
0.45 |
correct |
right |
female |
correct_right |
13.5939 |
0.5083 |
succesful_stop |
0.2 |
n/a |
n/a |
n/a |
female |
n/a |
17.1021 |
0.5083 |
unsuccesful_stop |
0.25 |
0.633 |
correct |
left |
male |
correct_left |
21.6103 |
0.5083 |
go |
n/a |
0.443 |
correct |
left |
male |
correct_left |
In this example, remap_columns combines the values from columns response_accuracy and
response_hand to produce a new column called response_type that specifies both response hand and correctness information using a single code.
Remove columns¶
Sometimes columns are added during intermediate processing steps. The remove_columns operation is useful for cleaning up unnecessary columns after these processing steps complete.
Remove columns parameters¶
Parameters for the remove_columns operation.
Parameter |
Type |
Description |
|---|---|---|
column_names |
list of str |
A list of columns to remove. |
ignore_missing |
boolean |
If true, missing columns are ignored, otherwise raise |
If one of the specified columns is not in the file and the ignore_missing
parameter is false, a KeyError is raised for the missing column.
Remove columns example¶
The following example specifies that the remove_columns operation should remove the stop_signal_delay,
response_accuracy, and face columns from the tabular data.
A JSON file with a single remove_columns transformation operation.
[{
"operation": "remove_columns",
"description": "Remove extra columns before the next step.",
"parameters": {
"column_names": ["stop_signal_delay", "response_accuracy", "face"],
"ignore_missing": true
}
}]
The results of executing this operation on the
sample remodel event file
are shown below.
The face column is not in the data, but it is ignored, since ignore_missing is true.
If ignore_missing had been false, a KeyError would have been raised.
Results of executing the remove_columns.
onset |
duration |
trial_type |
response_time |
response_hand |
sex |
|---|---|---|---|---|---|
0.0776 |
0.5083 |
go |
0.565 |
right |
female |
5.5774 |
0.5083 |
unsuccesful_stop |
0.49 |
right |
female |
9.5856 |
0.5084 |
go |
0.45 |
right |
female |
13.5939 |
0.5083 |
succesful_stop |
n/a |
n/a |
female |
17.1021 |
0.5083 |
unsuccesful_stop |
0.633 |
left |
male |
21.6103 |
0.5083 |
go |
0.443 |
left |
male |
Remove rows¶
The remove_rows operation eliminates rows in which the named column has one of the specified values.
This operation is useful for removing event markers corresponding to particular types of events
or, for example having n/a in a particular column.
Remove rows parameters¶
Parameters for remove_rows.
Parameter |
Type |
Description |
|---|---|---|
column_name |
str |
The name of the column to be tested. |
remove_values |
list |
A list of values to be tested for removal. |
The operation does not raise an error if a data file does not have a column named column_name or is missing a value in remove_values.
Remove rows example¶
The following remove_rows operation removes the rows whose trial_type column
contains either succesful_stop or unsuccesful_stop.
A JSON file with a single remove_rows transformation operation.
[{
"operation": "remove_rows",
"description": "Remove rows where trial_type is either succesful_stop or unsuccesful_stop.",
"parameters": {
"column_name": "trial_type",
"remove_values": ["succesful_stop", "unsuccesful_stop"]
}
}]
The results of executing the previous remove_rows operation on the sample remodel event file are:
The results of executing the previous remove_rows operation.
onset |
duration |
trial_type |
stop_signal_delay |
response_time |
response_accuracy |
response_hand |
sex |
|---|---|---|---|---|---|---|---|
0.0776 |
0.5083 |
go |
n/a |
0.565 |
correct |
right |
female |
9.5856 |
0.5084 |
go |
n/a |
0.45 |
correct |
right |
female |
21.6103 |
0.5083 |
go |
n/a |
0.443 |
correct |
left |
male |
After removing rows with trial_type equal to succesful_stop or unsuccesful_stop only the
three go trials remain.
Rename columns¶
The rename_columns operations uses a dictionary to map old column names into new ones.
Rename columns parameters¶
Parameters for rename_columns.
Parameter |
Type |
Description |
|---|---|---|
column_mapping |
dict |
The keys are the old column names and the values are the new names. |
ignore_missing |
bool |
If false, a |
If ignore_missing is false, a KeyError is raised if a column specified in
the mapping does not correspond to a column name in the data file.
Rename columns example¶
The following example renames the stop_signal_delay column to be stop_delay and
the response_hand to be hand_used.
A JSON file with a single rename_columns transformation operation.
[{
"operation": "rename_columns",
"description": "Rename columns to be more descriptive.",
"parameters": {
"column_mapping": {
"stop_signal_delay": "stop_delay",
"response_hand": "hand_used"
},
"ignore_missing": true
}
}]
The results of executing the previous rename_columns operation on the sample remodel event file are:
After the rename_columns operation is executed, the sample events file is:
onset |
duration |
trial_type |
stop_delay |
response_time |
response_accuracy |
hand_used |
sex |
|---|---|---|---|---|---|---|---|
0.0776 |
0.5083 |
go |
n/a |
0.565 |
correct |
right |
female |
5.5774 |
0.5083 |
unsuccesful_stop |
0.2 |
0.49 |
correct |
right |
female |
9.5856 |
0.5084 |
go |
n/a |
0.45 |
correct |
right |
female |
13.5939 |
0.5083 |
succesful_stop |
0.2 |
n/a |
n/a |
n/a |
female |
17.1021 |
0.5083 |
unsuccesful_stop |
0.25 |
0.633 |
correct |
left |
male |
21.6103 |
0.5083 |
go |
n/a |
0.443 |
correct |
left |
male |
Reorder columns¶
The reorder_columns operation reorders the indicated columns in the specified order. This operation is often used to place the most important columns near the beginning of the file for readability or to assure that all the data files in dataset have the same column order. Additional parameters control how non-specified columns are treated.
Reorder columns parameters¶
Parameters for the reorder_columns operation.
Parameter |
Type |
Description |
|---|---|---|
column_order |
list |
A list of columns in the order they should appear in the data. |
ignore_missing |
bool |
Controls handling column names in the reorder list that aren’t in the data. |
keep_others |
bool |
Controls handling of columns not in the reorder list. |
If ignore_missing is true and items in the reorder list do not exist in the file, the missing columns are ignored. On the other hand, if ignore_missing is false, a column name in the reorder list that is missing from the data raises a ValueError.
The keep_others parameter controls whether columns in the data that do not appear in the column_order list are dropped (keep_others is false) or put at the end in the relative order that they appear in the file (keep_others is true).
BIDS event files are required to have onset and duration as the first and second columns, respectively.
Reorder columns example¶
The reorder_columns operation in the following example specifies that the first four
columns of the dataset should be: onset, duration, response_time, and trial_type.
Since keep_others is false, these will be the only columns retained.
A JSON file with a single reorder_columns transformation operation.
[{
"operation": "reorder_columns",
"description": "Reorder columns.",
"parameters": {
"column_order": ["onset", "duration", "response_time", "trial_type"],
"ignore_missing": true,
"keep_others": false
}
}]
The results of executing the previous reorder_columns transformation on the sample remodel event file are:
Results of reorder_columns.
onset |
duration |
response_time |
trial_type |
|---|---|---|---|
0.0776 |
0.5083 |
0.565 |
go |
5.5774 |
0.5083 |
0.49 |
unsuccesful_stop |
9.5856 |
0.5084 |
0.45 |
go |
13.5939 |
0.5083 |
n/a |
succesful_stop |
17.1021 |
0.5083 |
0.633 |
unsuccesful_stop |
21.6103 |
0.5083 |
0.443 |
go |
Split rows¶
The split_rows operation is often used to convert event files from trial-level encoding to event-level encoding.
In trial-level encoding, all the events in a single trial (usually some variation of the cue-stimulus-response-feedback-ready sequence) are represented by a single row in the data file. Often, the onset corresponds to the presentation of the stimulus, and the other events are not reported or are implicitly reported.
In event-level encoding, each row represents the temporal marker for a single event. In this case a trial consists of a sequence of multiple events.
Split rows parameters¶
Parameters for the split_rows operation.
Parameter |
Type |
Description |
|---|---|---|
anchor_column |
str |
The name of the column that will be used for split_rows codes. |
new_events |
dict |
Dictionary whose keys are the codes to be inserted as new events |
remove_parent_event |
bool |
If true, remove parent event. |
The split_rows operation requires an anchor_column, which could be an existing column or a new column to be appended to the data. The purpose of the anchor_column is to hold the codes for the new events.
The new_events dictionary has the new events to be created. The keys are the new event codes to be inserted into the anchor_column. The values in new_events are themselves dictionaries. Each of these dictionaries has three keys:
onset_source is a list of items to be added to the onset of the event row being split to produce the
onsetcolumn value for the new event. These items can be any combination of numerical values and column names.duration a list of numerical values and/or column names whose values are to be added to compute the
durationcolumn value for the new event.copy_columns a list of column names whose values should be copied into each new event. Unlisted columns are filled with
n/a.
The split_rows operation sorts the split rows by the onset column and raises a TypeError
if the onset and duration are improperly defined.
The onset column is converted to numeric values as part splitting process.
Split rows example¶
The split_rows operation in the following example specifies that new rows should be added
to encode the response and stop signal. The anchor column is trial_type.
A JSON file with a single split_rows transformation operation.
[{
"operation": "split_rows",
"description": "add response events to the trials.",
"parameters": {
"anchor_column": "trial_type",
"new_events": {
"response": {
"onset_source": ["response_time"],
"duration": [0],
"copy_columns": ["response_accuracy", "response_hand", "sex", "trial_number"]
},
"stop_signal": {
"onset_source": ["stop_signal_delay"],
"duration": [0.5],
"copy_columns": ["trial_number"]
}
},
"remove_parent_event": false
}
}]
The results of executing this split_rows operation on the sample remodel event file are:
Results of the previous split_rows operation.
onset |
duration |
trial_type |
stop_signal_delay |
response_time |
response_accuracy |
response_hand |
sex |
|---|---|---|---|---|---|---|---|
0.0776 |
0.5083 |
go |
n/a |
0.565 |
correct |
right |
female |
0.6426 |
0 |
response |
n/a |
n/a |
correct |
right |
female |
5.5774 |
0.5083 |
unsuccesful_stop |
0.2 |
0.49 |
correct |
right |
female |
5.7774 |
0.5 |
stop_signal |
n/a |
n/a |
n/a |
n/a |
n/a |
6.0674 |
0 |
response |
n/a |
n/a |
correct |
right |
female |
9.5856 |
0.5084 |
go |
n/a |
0.45 |
correct |
right |
female |
10.0356 |
0 |
response |
n/a |
n/a |
correct |
right |
female |
13.5939 |
0.5083 |
succesful_stop |
0.2 |
n/a |
n/a |
n/a |
female |
13.7939 |
0.5 |
stop_signal |
n/a |
n/a |
n/a |
n/a |
n/a |
17.1021 |
0.5083 |
unsuccesful_stop |
0.25 |
0.633 |
correct |
left |
male |
17.3521 |
0.5 |
stop_signal |
n/a |
n/a |
n/a |
n/a |
n/a |
17.7351 |
0 |
response |
n/a |
n/a |
correct |
left |
male |
21.6103 |
0.5083 |
go |
n/a |
0.443 |
correct |
left |
male |
22.0533 |
0 |
response |
n/a |
n/a |
correct |
left |
male |
In a full processing example, it might make sense to rename trial_type to be
event_type and to delete the response_time and the stop_signal_delay columns,
since these items have been unfolded into separate events.
This could be accomplished in subsequent clean-up operations.
Remodel summarizations¶
Summarizations differ transformations in two respects: they do not modify the input data file, and they keep information about the results from each file that has been processed. Summarization operations may be used at several points in the operation list as checkpoints during debugging as well as for their more typical informational uses.
All summary operations have two required parameters: summary_name and summary_filename.
The summary_name is the unique key used to identify the particular incarnation of this summary in the dispatcher. Care should be taken to make sure that the summary_name is unique within a given JSON remodeling file if the same summary operation is used more than once within the file (e.g. for before and after summary information).
The summary_filename should also be unique and is used for saving the summary upon request.
When the remodeler is applied to full datasets rather than single files,
the summaries are saved in the derivatives/remodel/summaries directory under the dataset root.
A time stamp and file extension are appended to the summary_filename when the
summary is saved.
Summarize column names¶
The summarize_column_names tracks the unique column name patterns found in data files across the dataset and which files have these column names. This summary is useful for determining whether there are any non-conforming data files.
Often event files associated with different tasks have different column names, and this summary can be used to verify that the files corresponding to the same task have the same column names.
A more problematic issue is when some event files for the same task have reordered column names or use different column names.
Summarize column names parameters¶
The summarize_column_names operation has no parameters and only requires the summary_name and the summary_filename to specify the operation.
The summarize_column_names operation only has the two parameters required of all summaries.
Parameters for the summarize_column_names operation.
Parameter |
Type |
Description |
|---|---|---|
summary_name |
str |
A unique name used to identify this summary. |
summary_filename |
str |
A unique file basename to use for saving this summary. |
append_timecode |
bool |
(Optional) If True, append a time code to filename. |
Summarize column names example¶
The following example remodeling file produces a summary, which when saved
will appear with file name AOMIC_column_names_xxx.txt or
AOMIC_column_names_xxx.json where xxx is a timestamp.
A JSON file with a single summarize_column_names summarization operation.
[{
"operation": "summarize_column_names",
"description": "Summarize column names.",
"parameters": {
"summary_name": "AOMIC_column_names",
"summary_filename": "AOMIC_column_names"
}
}]
When this operation is applied to the sample remodel event file, the following text summary is produced.
Result of applying summarize_column_names to the sample remodel file.
Summary name: AOMIC_column_names
Summary type: column_names
Summary filename: AOMIC_column_names
Summary details:
Dataset: Number of files=1
Columns: ['onset', 'duration', 'trial_type', 'stop_signal_delay', 'response_time', 'response_accuracy', 'response_hand', 'sex']
sub-0013_task-stopsignal_acq-seq_events.tsv
Individual files:
sub-0013_task-stopsignal_acq-seq_events.tsv:
['onset', 'duration', 'trial_type', 'stop_signal_delay', 'response_time', 'response_accuracy', 'response_hand', 'sex']
Since we are only summarizing one event file, there is only one unique pattern – corresponding to the columns: onset, duration, trial_type, stop_signal_delay, response_time, response_accuracy, response_hand, and response_time.
When the dataset has multiple column name patterns, the summary lists unique pattern separately along with the names of the data files that have this pattern.
The JSON version of the summary is useful for programmatic manipulation, while the text version shown above is more readable.
Summarize column values¶
The summarize column values operation provides a summary of the number of times various column values appear in event files across the dataset.
Summarize column values parameters¶
The following table lists the parameters required for using the summary.
Parameters for the summarize_column_values operation.
Parameter |
Type |
Description |
|---|---|---|
summary_name |
str |
A unique name used to identify this summary. |
summary_filename |
str |
A unique file basename to use for saving this summary. |
skip_columns |
list |
A list of column names to omit from the summary. |
value_columns |
list |
A list of columns to omit the listing unique values. |
append_timecode |
bool |
(Optional) If True, append a time code to filename. |
max_categorical |
int |
(Optional) If given, the text summary shows top max_categorical values. |
values_per_line |
bool |
(Optional) If given, the text summary displays this |
In addition to the standard parameters, summary_name and summary_filename required of all summaries,
the summarize_column_values operation requires two additional lists to be supplied.
The skip_columns list specifies the names of columns to skip entirely in the summary.
Typically, the onset, duration, and sample columns are skipped, since they have unique values for
each row and their values have limited information.
The summarize_column_values is mainly meant for creating summary information about columns containing a finite number of distinct values. Columns that contain numeric information will usually have distinct entries for each row in a tabular file and are not amenable to such summarization. These columns could be specified as skip_columns, but another option is to designate them as value_columns. The value_columns are reported in the summary, but their distinct values are not reported individually.
For datasets that include multiple tasks, the event values for each task may be distinct.
The summarize_column_values operation does not separate by task, but expects the
calling programs filter the files by task as desired.
The run_remodel program supports selecting files corresponding to a particular task.
Two additional optional parameters are available for specifying aspects of the text summary output. The max_categorical optional parameter specifies how many unique values should be displayed for each column. The values_per_line controls how many categorical column values (with counts) are displayed on each line of the output. By default, 5 values are displayed.
Summarize column values example¶
The following example shows the JSON for including this operation in a remodeling file.
A JSON file with a single summarize_column_values summarization operation.
[{
"operation": "summarize_column_values",
"description": "Summarize the column values in an excerpt.",
"parameters": {
"summary_name": "AOMIC_column_values",
"summary_filename": "AOMIC_column_values",
"skip_columns": ["onset", "duration"],
"value_columns": ["response_time", "stop_signal_delay"]
}
}]
A text format summary of the results of executing this operation on the sample remodel event file is shown in the following example.
Sample summarize_column_values operation results in text format.
Summary name: AOMIC_column_values
Summary type: column_values
Summary filename: AOMIC_column_values
Overall summary:
Dataset: Total events=6 Total files=1
Categorical column values[Events, Files]:
response_accuracy:
correct[5, 1] n/a[1, 1]
response_hand:
left[2, 1] n/a[1, 1] right[3, 1]
sex:
female[4, 1] male[2, 1]
trial_type:
go[3, 1] succesful_stop[1, 1] unsuccesful_stop[2, 1]
Value columns[Events, Files]:
response_time[6, 1]
stop_signal_delay[6, 1]
Individual files:
sub-0013_task-stopsignal_acq-seq_events.tsv:
Total events=200
Categorical column values[Events, Files]:
response_accuracy:
correct[5, 1] n/a[1, 1]
response_hand:
left[2, 1] n/a[1, 1] right[3, 1]
sex:
female[4, 1] male[2, 1]
trial_type:
go[3, 1] succesful_stop[1, 1] unsuccesful_stop[2, 1]
Value columns[Events, Files]:
response_time[6, 1]
stop_signal_delay[6, 1]
Because the sample remodel event file only has 6 events, we expect that no value will be represented in more than 6 events. The column names corresponding to value columns just have the event counts in them.
This command was executed with the -i option in run_remodel,
results from the individual data files are shown after the overall summary.
The individual results are similar to the overall summary because only one data file
was processed.
For a more extensive example see the text and JSON format summaries of the sample dataset ds003645s_hed using the summarize_columns_rmdl.json remodeling file.
Summarize definitions¶
The summarize definitions operation provides a summary of the Def-expand tags found across the dataset,
nothing any ambiguous or erroneous ones. If working on a BIDS dataset, it will initialize with the known definitions
from the sidecar, reporting any deviations from the known definitions as errors.
Summarize definitions parameters¶
NOTE: This summary is still under development The following table lists the parameters required for using the summary.
Parameters for the summarize_definitions operation.
Parameter |
Type |
Description |
|---|---|---|
summary_name |
str |
A unique name used to identify this summary. |
summary_filename |
str |
A unique file basename to use for saving this summary. |
append_timecode |
bool |
(Optional) If True, append a time code to filename. |
The summarize_definitions is mainly meant for verifying consistency in unknown Def-expand tags. This comes up where you have an assembled dataset, but no longer have the definitions stored (or never created them to begin with).
Summarize definitions example¶
The following example shows the JSON for including this operation in a remodeling file.
A JSON file with a single summarize_definitions summarization operation.
[{
"operation": "summarize_definitions",
"description": "Summarize the definitions used in this dataset.",
"parameters": {
"summary_name": "HED_column_definition_summary",
"summary_filename": "HED_column_definition_summary"
}
}]
A text format summary of the results of executing this operation on the sub-003_task-FacePerception_run-3_events.tsv file of the eeg_ds_003645s_hed_column dataset is shown in the following example.
Sample summarize_definitions operation results in text format.
Summary name: HED_column_definition_summary
Summary type: definitions
Summary filename: HED_column_definition_summary
Overall summary:
Known Definitions: 17 items
cross-only: 2 items
description: A white fixation cross on a black background in the center of the screen.
contents: (Visual-presentation,(Background-view,Black),(Foreground-view,(Center-of,Computer-screen),(Cross,White)))
face-image: 2 items
description: A happy or neutral face in frontal or three-quarters frontal pose with long hair cropped presented as an achromatic foreground image on a black background with a white fixation cross superposed.
contents: (Visual-presentation,(Background-view,Black),(Foreground-view,((Center-of,Computer-screen),(Cross,White)),(Grayscale,(Face,Hair,Image))))
circle-only: 2 items
description: A white circle on a black background in the center of the screen.
contents: (Visual-presentation,(Background-view,Black),(Foreground-view,((Center-of,Computer-screen),(Circle,White))))
press-left-finger: 2 items
description: The participant presses a key with the left index finger to indicate a face symmetry judgment.
contents: ((Index-finger,(Experiment-participant,Left-side-of)),(Keyboard-key,Press))
press-right-finger: 2 items
description: The participant presses a key with the right index finger to indicate a face symmetry evaluation.
contents: ((Index-finger,(Experiment-participant,Right-side-of)),(Keyboard-key,Press))
famous-face-cond: 2 items
description: A face that should be recognized by the participants
contents: (Condition-variable/Face-type,(Image,(Face,Famous)))
unfamiliar-face-cond: 2 items
description: A face that should not be recognized by the participants.
contents: (Condition-variable/Face-type,(Image,(Face,Unfamiliar)))
scrambled-face-cond: 2 items
description: A scrambled face image generated by taking face 2D FFT.
contents: (Condition-variable/Face-type,(Image,(Disordered,Face)))
first-show-cond: 2 items
description: Factor level indicating the first display of this face.
contents: ((Condition-variable/Repetition-type,Item-interval/0,(Face,Item-count/1)))
immediate-repeat-cond: 2 items
description: Factor level indicating this face was the same as previous one.
contents: ((Condition-variable/Repetition-type,Item-interval/1,(Face,Item-count/2)))
delayed-repeat-cond: 2 items
description: Factor level indicating face was seen 5 to 15 trials ago.
contents: (Condition-variable/Repetition-type,(Face,Item-count/2),(Item-interval,(Greater-than-or-equal-to,Item-interval/5)))
left-sym-cond: 2 items
description: Left index finger key press indicates a face with above average symmetry.
contents: (Condition-variable/Key-assignment,((Asymmetrical,Behavioral-evidence),(Index-finger,(Experiment-participant,Right-side-of))),((Behavioral-evidence,Symmetrical),(Index-finger,(Experiment-participant,Left-side-of))))
right-sym-cond: 2 items
description: Right index finger key press indicates a face with above average symmetry.
contents: (Condition-variable/Key-assignment,((Asymmetrical,Behavioral-evidence),(Index-finger,(Experiment-participant,Left-side-of))),((Behavioral-evidence,Symmetrical),(Index-finger,(Experiment-participant,Right-side-of))))
face-symmetry-evaluation-task: 2 items
description: Evaluate degree of image symmetry and respond with key press evaluation.
contents: (Experiment-participant,Task,(Discriminate,(Face,Symmetrical)),(Face,See),(Keyboard-key,Press))
blink-inhibition-task: 2 items
description: Do not blink while the face image is displayed.
contents: (Experiment-participant,Inhibit-blinks,Task)
fixation-task: 2 items
description: Fixate on the cross at the screen center.
contents: (Experiment-participant,Task,(Cross,Fixate))
initialize-recording: 2 items
description:
contents: (Recording)
Ambiguous Definitions: 0 items
Errors: 0 items
Since this file didn’t have any ambiguous or incorrect Def-expand groups, those sections are empty.
Ambiguous definitions are those that take a placeholder, but it doesn’t have enough information
to be sure to which tag the placeholder applies.
Erroneous ones are ones with conflicting expanded forms.
Currently, summaries are not generated for individual files, but this is likely to change in the future.
Below is a simple example showing the format when erroneous or ambiguous definitions are found.
Sample input for summarize_definitions operation documenting ambiguous/erroneous definitions.
((Def-expand/Initialize-recording,(Recording)),Onset)
((Def-expand/Initialize-recording,(Recording, Event)),Onset)
(Def-expand/Specify-age/1,(Age/1, Item-count/1))
Sample summarize_definitions operation error results in text format.
Summary name: HED_column_definition_summary
Summary type: definitions
Summary filename: HED_column_definition_summary
Overall summary:
Known Definitions: 1 items
initialize-recording: 2 items
description:
contents: (Recording)
Ambiguous Definitions: 1 items
specify-age/#: (Age/#,Item-count/#)
Errors: 1 items
initialize-recording:
(Event,Recording)
It is assumed the first definition encountered is the correct definition, unless the first one is ambiguous.
Thus, it finds (Def-expand/Initialize-recording,(Recording) and considers it valid, before encountering
(Def-expand/Initialize-recording,(Recording, Event)), which is now deemed an error.
Summarize HED type¶
The summarize_hed_type operation is designed to extract experimental design matrices or other experimental structure. This summary operation assumes that the structure in question is suitably annotated with HED (Hierarchical Event Descriptors). The HED conditions and design matrices explains how this works.
Summarize HED type parameters¶
The summarize_hed_type operation provides detailed information about a specified tag,
usually Condition-variable or Task.
This summary provides useful information about experimental design.
Parameters for the summarize_hed_type operation.
Parameter |
Type |
Description |
|---|---|---|
summary_name |
str |
A unique name used to identify this summary. |
summary_filename |
str |
A unique file basename to use for saving this summary. |
type_tag |
str |
Tag to produce a summary for (most often condition-variable). |
append_timecode |
bool |
(Optional) If True, append a time code to filename. |
In addition to the two standard parameters (summary_name and summary_filename), the type_tag parameter is required. Only one tag can be given, so you must provide a separate operations in the remodel file for multiple type tags.
Summarize HED type example¶
A JSON file with a single summarize_hed_type summarization operation.
[{
"operation": "summarize_hed_type",
"description": "Summarize column names.",
"parameters": {
"summary_name": "AOMIC_condition_variables",
"summary_filename": "AOMIC_condition_variables",
"type_tag": "condition-variable"
}
}]
The results of executing this operation on the sample remodel event file are shown below.
Text summary of summarize_hed_types operation on the sample remodel file.
Summary name: AOMIC_condition_variables
Summary type: hed_type_summary
Summary filename: AOMIC_condition_variables
Overall summary:
Dataset: Type=condition-variable Type values=1 Total events=6 Total files=1
image-sex: 2 levels in 6 event(s)s out of 6 total events in 1 file(s)
female-image-cond [4,1]: ['Female', 'Image', 'Face']
male-image-cond [2,1]: ['Male', 'Image', 'Face']
Individual files:
aomic_sub-0013_excerpt_events.tsv:
Type=condition-variable Total events=6
image-sex: 2 levels in 6 events
female-image-cond [4 events, 1 files]:
Tags: ['Female', 'Image', 'Face']
male-image-cond [2 events, 1 files]:
Tags: ['Male', 'Image', 'Face']
Because summarize_hed_type is a HED operation,
a HED schema version is required and a JSON sidecar is also usually needed.
This summary was produced by using hed_version="8.1.0" when creating the dispatcher
and using the sample remodel sidecar file in the do_op.
The sidecar provides the annotations that use the condition-variable tag in the summary.
For a more extensive example, see the text and JSON format summaries of the sample dataset ds003645s_hed using the summarize_hed_types_rmdl.json remodeling file.
Summarize HED validation¶
The summarize_hed_validation operation runs the HED validator on the requested data and produces a summary of the errors. See the HED validation guide for available methods of running the HED validator.
Summarize HED validation parameters¶
In addition to the required summary_name and summary_filename parameters, the summarize_hed_validation operation has a required boolean parameter check_for_warnings. If check_for_warnings is false, the summary will not report warnings.
Parameters for the summarize_hed_validation operation.
Parameter |
Type |
Description |
|---|---|---|
summary_name |
str |
A unique name used to identify this summary. |
summary_filename |
str |
A unique file basename to use for saving this summary. |
append_timecode |
bool |
(Optional) If True, append a time code to filename. |
check_for_warnings |
bool |
(Optional) If true, warnings are reported in addition to errors. |
The summarize_hed_validation is a HED operation and the calling program must provide a HED schema version and usually a JSON sidecar containing the HED annotations.
The validation process takes place in two stages: first the JSON sidecar is validated. This strategy is used because a single error in the JSON sidecar can generate an error message for every line in the corresponding data file.
If the JSON sidecar has errors (warnings don’t count), the validation process is terminated without validation of the data file and assembled HED annotations.
If the JSON sidecar does not have errors, the validator assembles the annotations for each line in the data files and validates the assembled HED annotation. Data file-wide consistency, such as matched onsets and offsets, is also checked.
Summarize HED validation example¶
A JSON file with a single summarize_hed_validation summarization operation.
[{
"operation": "summarize_hed_validation",
"description": "Summarize validation errors in the sample dataset.",
"parameters": {
"summary_name": "AOMIC_sample_validation",
"summary_filename": "AOMIC_sample_validation",
"check_for_warnings": true
}
}]
To demonstrate the output of the validation operation, we modified the first row of the
sample remodel event file
so that trial_type column contained the value baloney rather than go.
This modification generates a warning because the meaning of baloney is not defined
in the sample remodel sidecar file.
The results of executing the example operation with the modified file are shown
in the following example.
Text summary of summarize_hed_validation operation on a modified sample data file.
Summary name: AOMIC_sample_validation
Summary type: hed_validation
Summary filename: AOMIC_sample_validation
Summary details:
Dataset: [1 sidecar files, 1 event files]
task-stopsignal_acq-seq_events.json: 0 issues
sub-0013_task-stopsignal_acq-seq_events.tsv: 6 issues
Individual files:
sub-0013_task-stopsignal_acq-seq_events.tsv: 1 sidecar files
task-stopsignal_acq-seq_events.json has no issues
sub-0013_task-stopsignal_acq-seq_events.tsv issues:
HED_UNKNOWN_COLUMN: WARNING: Column named 'onset' found in file, but not specified as a tag column or identified in sidecars.
HED_UNKNOWN_COLUMN: WARNING: Column named 'duration' found in file, but not specified as a tag column or identified in sidecars.
HED_UNKNOWN_COLUMN: WARNING: Column named 'response_time' found in file, but not specified as a tag column or identified in sidecars.
HED_UNKNOWN_COLUMN: WARNING: Column named 'response_accuracy' found in file, but not specified as a tag column or identified in sidecars.
HED_UNKNOWN_COLUMN: WARNING: Column named 'response_hand' found in file, but not specified as a tag column or identified in sidecars.
HED_SIDECAR_KEY_MISSING[row=0,column=2]: WARNING: Category key 'baloney' does not exist in column. Valid keys are: ['succesful_stop', 'unsuccesful_stop', 'go']
This summary was produced using HED schema version hed_version="8.1.0" when creating the dispatcher
and using the sample remodel sidecar file in the do_op.
Summarize sidecar from events¶
The summarize sidecar from events operation generates a sidecar template from the event files in the dataset.
Summarize sidecar from events parameters¶
The following table lists the parameters required for using the summary.
Parameters for the summarize_sidcar_from_events operation.
Parameter |
Type |
Description |
|---|---|---|
summary_name |
str |
A unique name used to identify this summary. |
summary_filename |
str |
A unique file basename to use for saving this summary. |
skip_columns |
list |
A list of column names to omit from the sidecar. |
value_columns |
list |
A list of columns to treat as value columns in the sidecar. |
append_timecode |
bool |
(Optional) If True, append a time code to filename. |
The standard summary parameters, summary_name and summary_filename are required. The summary_name is the unique key used to identify the particular incarnation of this summary in the dispatcher. Since a particular operation file may use a given operation multiple times, care should be taken to make sure that it is unique.
The summary_filename should also be unique and is used for saving the summary upon request.
When the remodeler is applied to full datasets rather than single files,
the summaries are saved in the derivatives/remodel/summaries directory under the dataset root.
A time stamp and file extension are appended to the summary_filename when the
summary is saved.
In addition to the standard parameters, summary_name and summary_filename required of all summaries, the summarize_sidecar_from_events operation requires two additional lists to be supplied. The skip_columns list specifies the names of columns to skip entirely in generating the sidecar template. The value_columns list specifies the names of columns to treat as value columns when generating the sidecar template.
Summarize sidecar from events example¶
The following example shows the JSON for including this operation in a remodeling file.
A JSON file with a single summarize_sidecar_from_events summarization operation.
[{
"operation": "summarize_sidecar_from_events",
"description": "Generate a sidecar from the excerpted events file.",
"parameters": {
"summary_name": "AOMIC_generate_sidecar",
"summary_filename": "AOMIC_generate_sidecar",
"skip_columns": ["onset", "duration"],
"value_columns": ["response_time", "stop_signal_delay"]
}
}]
The results of executing this operation on the sample remodel event file are shown in the following example using the text format.
Sample summarize_sidecar_from_events operation results in text format.
Summary name: AOMIC_generate_sidecar
Summary type: events_to_sidecar
Summary filename: AOMIC_generate_sidecar
Dataset: Currently no overall sidecar extraction is available
Individual files:
aomic_sub-0013_excerpt_events.tsv: Total events=6 Skip columns: ['onset', 'duration']
Sidecar:
{
"trial_type": {
"Description": "Description for trial_type",
"HED": {
"go": "(Label/trial_type, Label/go)",
"succesful_stop": "(Label/trial_type, Label/succesful_stop)",
"unsuccesful_stop": "(Label/trial_type, Label/unsuccesful_stop)"
},
"Levels": {
"go": "Here describe column value go of column trial_type",
"succesful_stop": "Here describe column value succesful_stop of column trial_type",
"unsuccesful_stop": "Here describe column value unsuccesful_stop of column trial_type"
}
},
"response_accuracy": {
"Description": "Description for response_accuracy",
"HED": {
"correct": "(Label/response_accuracy, Label/correct)"
},
"Levels": {
"correct": "Here describe column value correct of column response_accuracy"
}
},
"response_hand": {
"Description": "Description for response_hand",
"HED": {
"left": "(Label/response_hand, Label/left)",
"right": "(Label/response_hand, Label/right)"
},
"Levels": {
"left": "Here describe column value left of column response_hand",
"right": "Here describe column value right of column response_hand"
}
},
"sex": {
"Description": "Description for sex",
"HED": {
"female": "(Label/sex, Label/female)",
"male": "(Label/sex, Label/male)"
},
"Levels": {
"female": "Here describe column value female of column sex",
"male": "Here describe column value male of column sex"
}
},
"response_time": {
"Description": "Description for response_time",
"HED": "(Label/response_time, Label/#)"
},
"stop_signal_delay": {
"Description": "Description for stop_signal_delay",
"HED": "(Label/stop_signal_delay, Label/#)"
}
}
Remodel implementation¶
Operations are defined as classes that extent BaseOp regardless of whether
they are transformations or summaries. However, summaries must also implement
an additional supporting class that extends BaseSummary to hold the summary information.
In order to be executed by the remodeling functions,
an operation must appear in the valid_operations dictionary.
All operations must provide a PARAMS dictionary, a constructor that calls the
base class constructor, and a do_ops method.
The PARAMS dictionary¶
The class-wide PARAMS dictionary has operation, required_parameters and optional_parameters keys.
The required_parameters and optional_parameters have values that are themselves dictionaries
specifying the names and types of the operation parameters.
The following example shows the PARAMS dictionary for the RemoveColumnsOp class.
The class-wide PARAMS dictionary for the RemoveColumnsOp class.
PARAMS = {
"operation": "remove_columns",
"required_parameters": {
"column_names": list,
"ignore_missing": bool
},
"optional_parameters": {}
}
The PARAMS dictionary allows the remodeling tools to check the syntax of the remodel input file for errors.
Operation class constructor¶
All the operation classes have constructors that start with a call to the superclass constructor BaseOp.
The following example shows the constructor for the RemoveColumnsOp class.
The class-wide PARAMS dictionary for the RemoveColumnsOp class.
def __init__(self, parameters):
super().__init__(self.PARAMS, parameters)
self.column_names = parameters['column_names']
ignore_missing = parameters['ignore_missing']
if ignore_missing:
self.error_handling = 'ignore'
else:
self.error_handling = 'raise'
After the call to the base class constructor, the operation constructor assigns the operation-specific values to class properties and does any additional required operation-specific checks to assure that the parameters are valid.
The do_op implementation¶
The main method that must be implemented by each operation is do_op, which takes
an instance of the Dispatcher class as the first parameter and a Pandas DataFrame
representing the event file as the second parameter.
A third required parameter is a name used to identify the event file in error messages and summaries.
This name is usually the filename or the filepath from the dataset root.
An additional optional argument, a sidecar containing HED annotations,
only need be included for HED operations.
The following example shows a sample implementation for do_op.
The implementation of do_op for the RemoveColumnsOp class.
def do_op(self, dispatcher, df, name, sidecar=None):
return df.drop(self.remove_names, axis=1, errors=self.error_handling)
The do_op in this case is a wrapper for the underlying Pandas DataFrame
operation for removing columns.
IMPORTANT NOTE: The do_op operation always assumes that n/a values have been
replaced by numpy.NaN values in the incoming dataframe df.
The Dispatcher class has a static method prep_data that does this replacement.
At the end of running all the remodeling operations on a data file Dispatcher run_operations
method replaces all of the numpy.NaN values with n/a, the value expected by BIDS.
This operation is performed by the Dispatcher static method post_proc_data.
The do_op for summarization¶
The do_op operation for summarization operations has a slightly different form,
as it serves primarily as a wrapper for the actual summary information as illustrated
by the following example.
The implementation of do_op for SummarizeColumnNamesOp.
def do_op(self, dispatcher, df, name, sidecar=None):
summary = dispatcher.summary_dict.get(self.summary_name, None)
if not summary:
summary = ColumnNameSummary(self)
dispatcher.summary_dict[self.summary_name] = summary
summary.update_summary({"name": name, "column_names": list(df.columns)})
return df
A do_op operation for a summarization checks the dispatcher to see if the
summary name is already in the dispatcher’s summary_dict.
If that summary is not yet in the summary_dict,
the operation creates a BaseSummary object for its summary (e.g., ColumnNameSummary)
and adds this object to the dispatcher’s summary_dict,
otherwise the operation fetches the BaseSummary object from
It then asks its BaseSummary object to update the summary based on the dataframe
as explained in the next section.
Additional requirements for summarization¶
Any summary operation must implement a supporting class that extends BaseSummary.
This class is used to hold and accumulate the information specific to the summary.
This support class must implement two methods: update_summary and get_summary_details.
The update_summary method is called by its associated BaseOp operation during the do_op
to update the summary information based on the current dataframe.
The update_summary information takes a single parameter, which is a dictionary of information
specific to this operation.
The update_summary method required to be implemented by all BaseSummary objects.
def update_summary(self, summary_dict)
In the example do_op for ColumnNamesOp,
the dictionary is contains keys for name and `column_names.
The get_summary_details returns a dictionary with the summary-specific information
currently in the summary.
The BaseSummary provides universal methods for converting this summary to JSON or text format.
The get_summary_details method required to be implemented by all BaseSummary objects.
get_summary_details(self, verbose=True)
The operation associated with this instance of it associated with a given format implementation